About QC-bind
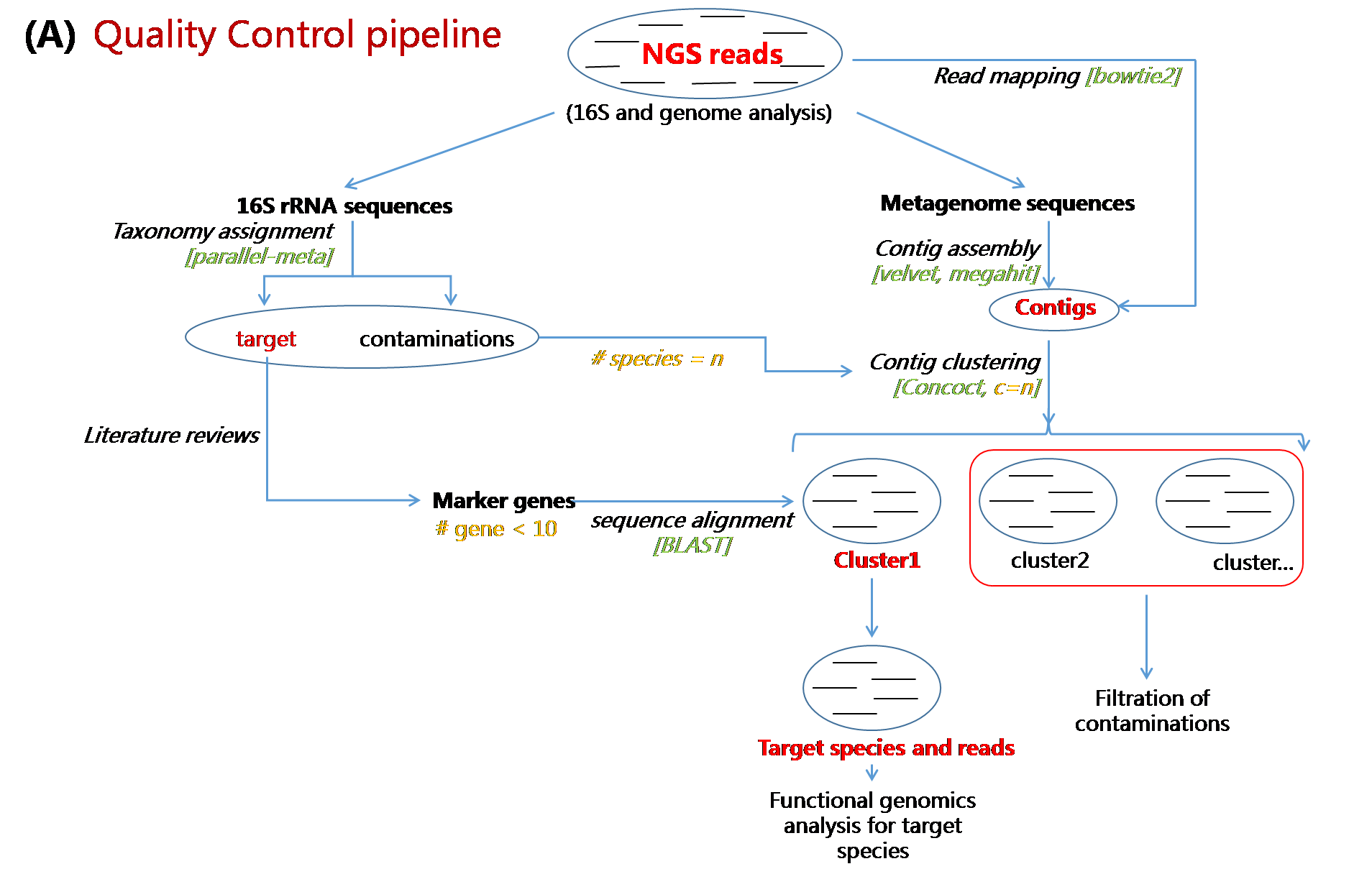
In this study, we have attempted to address these issues, by designing more accurate and flexible pipeline, QC-Blind, that could not only reduce the false positive rate, but also needs only a few marker genes for differentiating reads from target species and contaminations. We applied unsupervised methods to cluster pre-assembled contigs into species-level groups, and then utilized marker genes of target species to identify contig clusters that belong to certain groups. Extensive downstream evaluations have been performed to prove this pipeline to be highly accurate and speedy, for all of on in silico, ab initio and in vivo datasets. As most microbial contaminations could be removed and almost complete genomics information of target species could be preserved after its processing, this pipeline represents the frontier in NGS data quality control that progresses towards the solution for critical challenge of microbial contaminations in NGS data. Unlike traditional alignment-based method that highly depends on genomic information, QC-Blind pipeline represents the frontier in NGS data quality control. It precisely resolved sequence clusters for different species without any reference genome, and then identified target clusters with limited number of marker genes. The greatest advantage of our pipeline, QC-Blind, is its ability to perform quality control when neither target or contaminations has completegenomes24, where traditional methods fail. It’s an “Amplifier” that enlarged the power of marker genes to retrieve possibly the whole genome of target species.Another important feature of QC-Blind is that the selection of marker genes is flexible and context-dependent, thus providing a lot of room for improvement for its application. Thus the power of this approach is not limited to samples with single target species, or merely contamination removal.